|
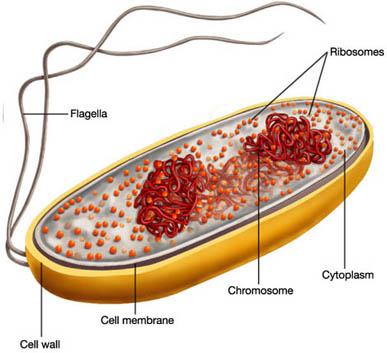 Bacterial chromosomal DNA is
not confined to an envelope-enclosed organelle such as the nucleus in eukaryotes,
yet the volume it occupies has to be reduced below that of the cell. An unconstrained
chromosomal DNA molecule of 1.6 mm as found in E. coli would form
a random coil with a volume of ~200 um3. Due to the action of
a number of factors a DNA molecule of this size can nevertheless be fit into
a cell that is only 2 um long and 1 um wide. These factors include macromolecular
crowding (yielding a phase separation between cytoplasm and DNA) and DNA
supercoiling.
Bacterial chromosomal DNA is
not confined to an envelope-enclosed organelle such as the nucleus in eukaryotes,
yet the volume it occupies has to be reduced below that of the cell. An unconstrained
chromosomal DNA molecule of 1.6 mm as found in E. coli would form
a random coil with a volume of ~200 um3. Due to the action of
a number of factors a DNA molecule of this size can nevertheless be fit into
a cell that is only 2 um long and 1 um wide. These factors include macromolecular
crowding (yielding a phase separation between cytoplasm and DNA) and DNA
supercoiling.
Our primary interest is the role that a group of small architectural
proteins (called nucleoid-associated proteins - NAP's) plays in organizing
and compacting the bacterial chromosomal DNA. These proteins can be divided
into two main groups based on their mode of action: bridgers and benders.
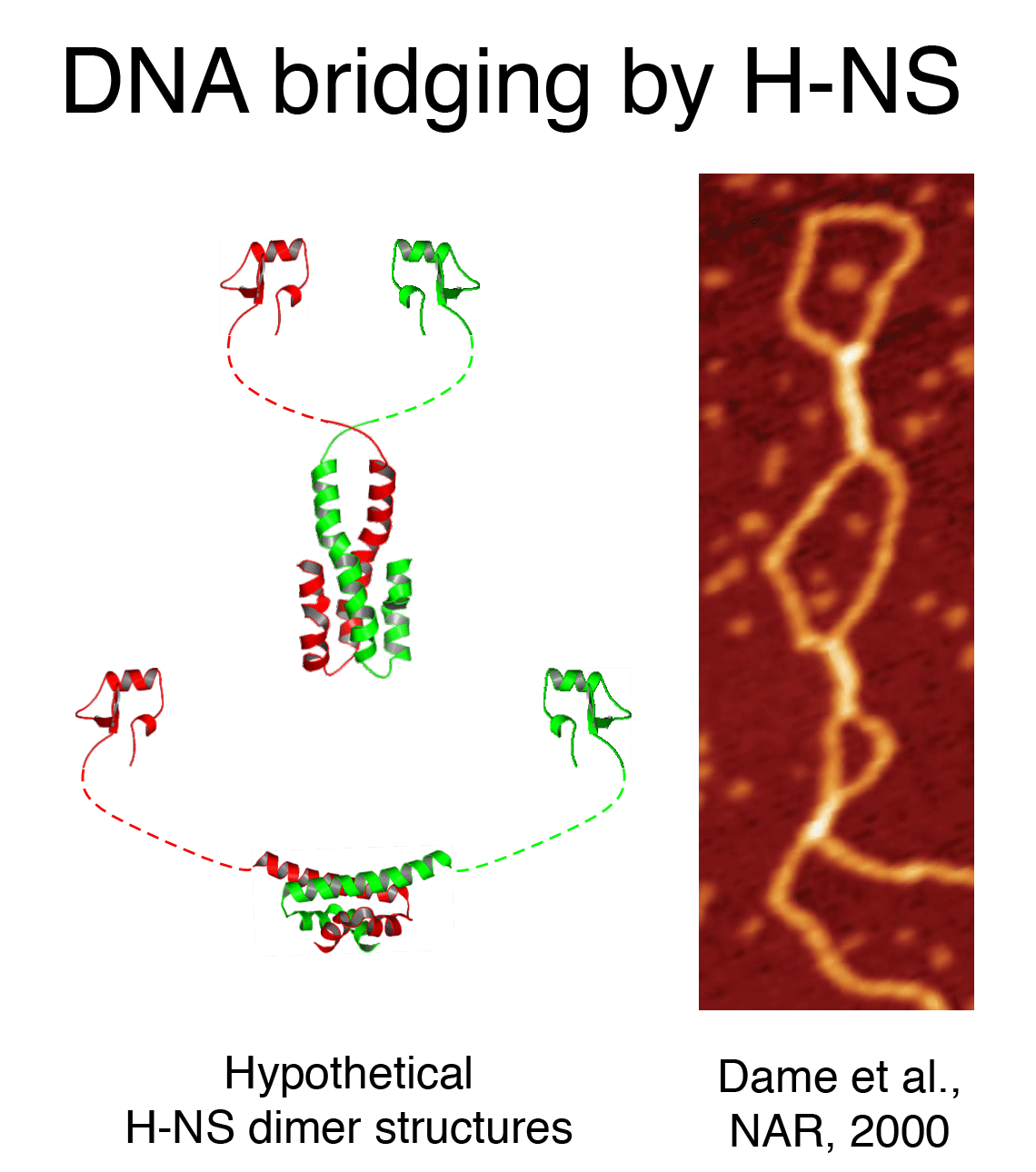 DNA bridging
proteins (H-NS, StpA, Lrp, SMC proteins, …) usually have a multimeric structure
with multiple DNA binding domains, providing a means to interact with two
DNA duplexes simultaneously. We have investigated the activity of H-NS like
proteins as organizers of bacterial chromatin and the structural basis of
repression mechanisms. SMC proteins are much larger in size when compared
to the other NAP's and presumable act by enclosing multiple DNA duplexes
rather than by directly interacting with the DNA.
DNA bridging
proteins (H-NS, StpA, Lrp, SMC proteins, …) usually have a multimeric structure
with multiple DNA binding domains, providing a means to interact with two
DNA duplexes simultaneously. We have investigated the activity of H-NS like
proteins as organizers of bacterial chromatin and the structural basis of
repression mechanisms. SMC proteins are much larger in size when compared
to the other NAP's and presumable act by enclosing multiple DNA duplexes
rather than by directly interacting with the DNA.
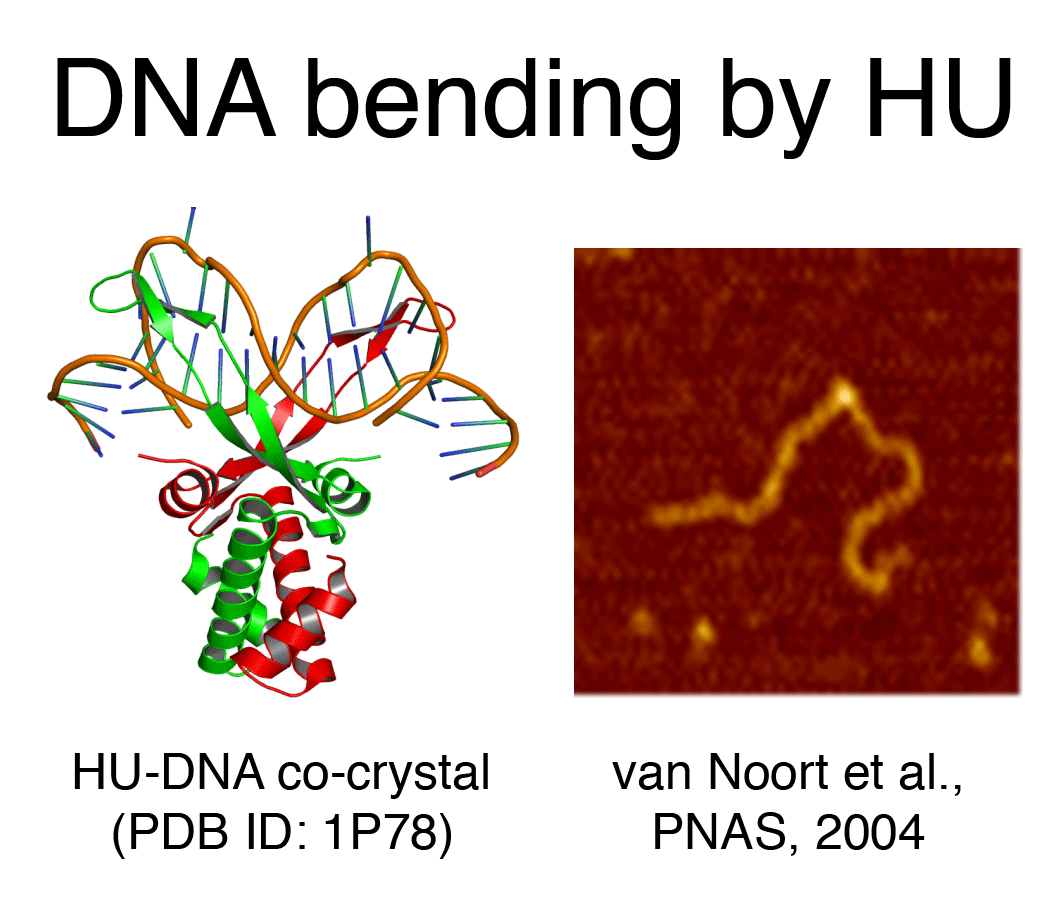 DNA bending proteins (HU, IHF, Fis, ... ) bind DNA either at specific or non-specific sites and in doing so distort it. The bending by these proteins amounts 50-90 (in the case of Fis) and up to ~180 degrees (in the case of HU and IHF). Both bridging and bending yield compaction and aid in the organization of the chromosomal DNA. Interestingly both HU and Fis have an alternative 'binding mode' in which they form filaments around DNA.
DNA bending proteins (HU, IHF, Fis, ... ) bind DNA either at specific or non-specific sites and in doing so distort it. The bending by these proteins amounts 50-90 (in the case of Fis) and up to ~180 degrees (in the case of HU and IHF). Both bridging and bending yield compaction and aid in the organization of the chromosomal DNA. Interestingly both HU and Fis have an alternative 'binding mode' in which they form filaments around DNA.
Most of these proteins do not just play
a role as organizers of bacterial chromatin, but also act as regulators of
transcription. For instance, IHF and Fis act as direct activators of transcription
by binding at specific sites upstream of the promoter. Similarly, H-NS directly
represses transcription by binding preferentially at A/T-rich/flexible regions
found close to H-NS sensitive promoters. Besides such direct effects, transcription
of a large set of genes is regulated indirectly by opposing the effects of
a second NAP. For instance, many genes repressed by H-NS, are specifically
de-repressed by IHF, HU or Fis. The latter set of proteins is expressed differently
during different phases of growth, which allows different subsets of genes
to be de-repressed. Since these proteins act both at specific sites and non-specific
sites, their differences in expression probably also give rise to differences
in global genome organization that indirectly affect transcription levels
from a substantial number of additional genes.
|



